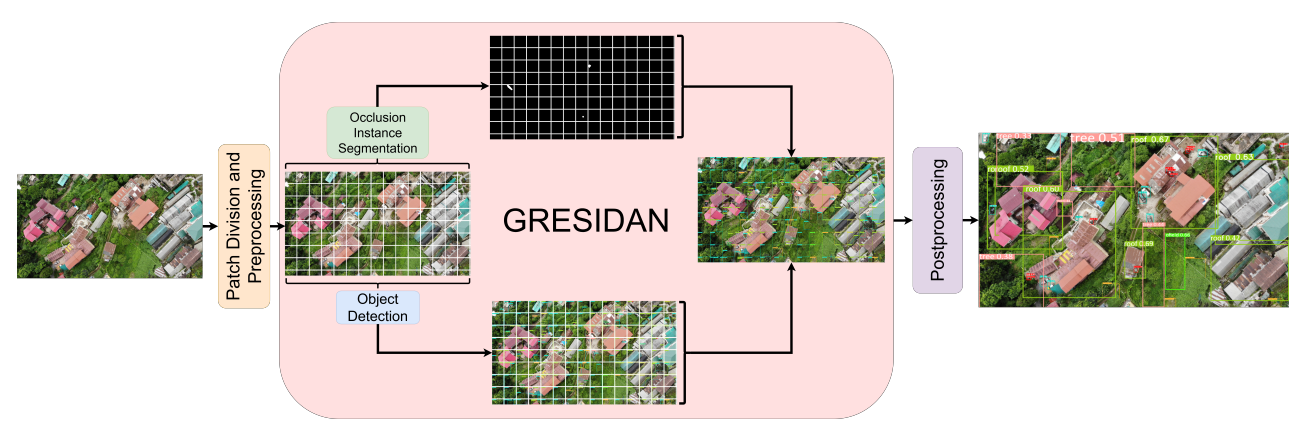
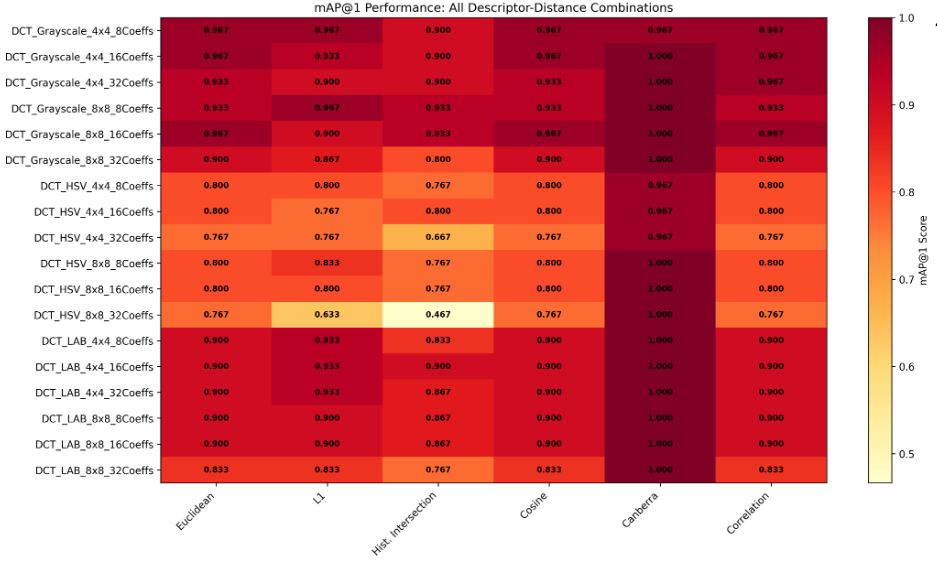
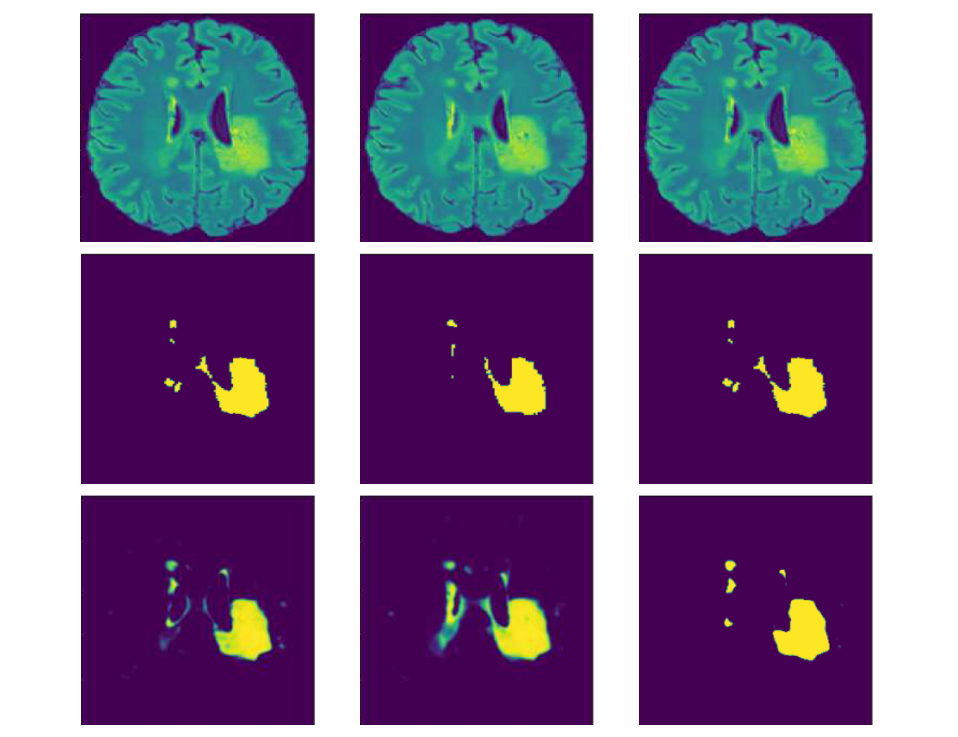
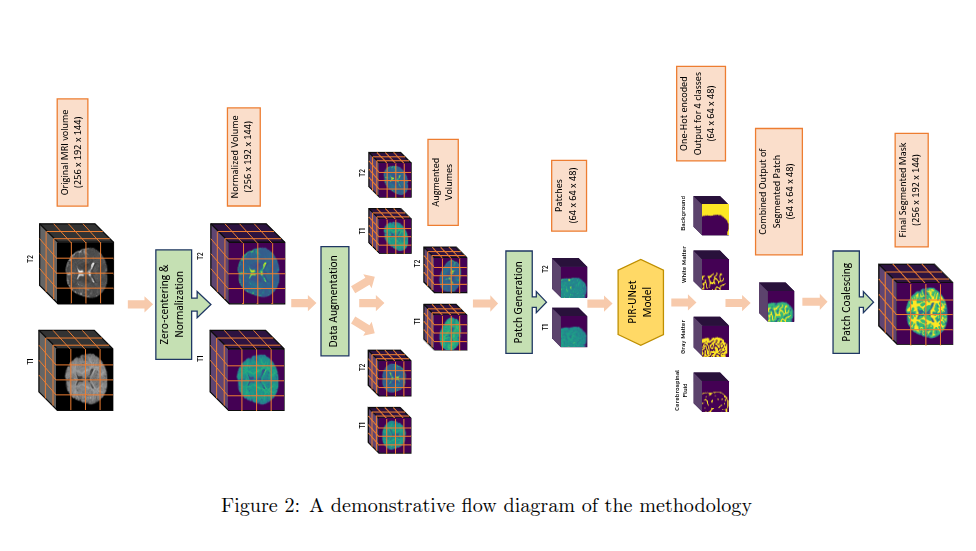
DocRevive: An End-to-End Framework for Document Text Restoration
In Document Analysis and Recognition, the challenge of reconstructing damaged, occluded, or incomplete text remains a critical
yet unexplored problem. Subsequent document understanding tasks can
benefit from a document reconstruction process
In Document Analysis and Recognition, the challenge of reconstructing damaged, occluded, or incomplete text remains a critical
yet unexplored problem. Subsequent document understanding tasks can
benefit from a document reconstruction process. This paper presents a
novel end-to-end pipeline designed to address this challenge, combining
state-of-the-art Optical Character Recognition (OCR), advanced image
analysis, and the contextual power of large language models (LLMs) and
diffusion-based models to restore and reconstruct text while preserving
visual integrity. We create a synthetic dataset simulating diverse document degradation scenarios, establishing a benchmark for restoration
tasks. Our pipeline detects and recognizes text, identifies degradation,
and uses LLMs for semantically coherent reconstruction. A diffusion-based module reintegrates text seamlessly, matching font, size, and alignment. To evaluate restoration quality, we propose a Unified Context Sim-
ilarity Metric (UCSM), incorporating edit, semantic, and length similarities with a contextual predictability measure that penalizes deviations
when the correct text is contextually obvious. Our work advances document restoration, benefiting archival research and digital preservation
while setting a new standard for text reconstruction.